Over the last few days I’ve started moving my web material to a new host. As a result I’ve decided to consolidate my blog there as well. So, I won’t be posting here anymore. The new blog is located at http://blog.rguha.net and carries over all the posts, tags and comments.
Browsing the latest articles in JCIM, I came across one by Sander et al that discussed the design of a drug discovery informatics system employed at Actelion. The main claim to fame of the work appears to be the fact that it was built from scratch and so is vendor independent.
While somewhat interesting, one question jumped out at me: what is the value of this paper? The system is specific to this one company so it’s not like I can access the code or the workflows. I can’t even buy this software. In this case, they do provide public access to some of their tools, so it’s not a totally “opaque” paper. But there are other examples where one cannot really even try out the tools described in the paper.
While I see the value of the paper from the authors point of view (spreading the word, publications being “currency”, etc.), such papers have always felt a little pointless to me, as a reader. What can I do after reading this paper? Is there anything I can follow-up on?
Posted in Literature | 13 Comments »
Today while working on a project I needed to get access to the Gene Ontology hierarchy. While there a number of GO browsers such as Amigo, I needed access to the raw data to generate a graph that I could then slice and dice. A few minutes with Python led to a simple solution.
The program parses the OBO 1.2 formatted GO data file (either by directly downloading it or from a local file) and outputs a flat dictionary listing the term ID’s, names, namespace etc and a network representation of the GO hierarchy in ncol format. It uses a simple (and relatively non-robust) class to represent the data as an undirected graph (not really correct), though it’d be easy to use something like igraph to start doing some real network analysis. It’s certainly not a comprehensive solution, but I thought I’d put it out there.
Posted in software | Tagged go, network, python | 3 Comments »
I’ve been working for some time with the PubChem Bioassay collection – a set of 1293 assays that cover a range of techniques (enzymatic, phenotypic etc.), targets and sizes (from 20 molecules to 200,000 molecules). In addition, some assays are primary, high-throughput assays whereas a number of them are smaller, confirmatory assays. While an extremely valuable collection, one of the drawbacks is the lack of curation. This has led to some people saying that the data is too noisy to be useful. Yes, the noise is a problem, but I think there’s still useful data to extract and model.
One of the problems that I have faced is that while one can perform a full text search for assays on PubChem, there is no form of annotations on the assays themselves. One effect of this is that it is difficult to link an assay to other biological resources (though for enzymatic assays, one can determine a Pubmed protein identifier). While working on my bioassay network project, I needed annotations and I didn’t want to do it manually.
Posted in cheminformatics, text mining, visualization | Tagged annotation, database, go, network, pubchem, text mining, visualization | 1 Comment »
Last year, John Van Drie and I published two papers (here and here) on the Structure Activity Landscape Index, (SALI) which is a way to view SAR data as a network of compounds. Along with the paper ,I put up a simple Java application (licensed under the LGPL) to generate and explore these networks. – you only need to provide a file containing SMILES and activities. It’s based on ZGRViewer – a very slick GUI for Graphviz generated networks. I finally got around to reorganizing the code and putting it up on a GitHub repository. You can get more details of the application and the last stable version here.
Posted in cheminformatics, software | Tagged graphviz, network, sali | Leave a Comment »
Using the model deployment and prediction service, I put up the two linear regression models I had built so far (described in more detail here) While REST is nice, a simple web page that allows you to paste a set of SMILES and get back predictions is handy. So I whipped together a simple interface to the prediction service, allowing one to select a model, view the author-generated description and a get a nice (sortable!) table of predicted values. View it here. As noted in my previous post it’s not going to be very fast, but hopefully that will change in the future.
Posted in software | Tagged ons, prediction, qsar, REST, solubility | Leave a Comment »
Over the past few days I’ve been developing some predictive models in R, for the solubility data being generated as part of the ONS Solubility Challenge. As I develop the models I put up a brief summary of the results on the wiki. In the end however, we’d like to use these models to predict the solubility of untested compounds. While anybody can send me a SMILES string and get back a prediction, it’s more useful (and less work for me!) if a user can do it themselves. This requires that the models be deployed and made available as a web page or a service. Last year I developed a series of statistical web services based on R. The services were written in Java and are described in this paper. Since I’m working more with REST services these days, I wanted to see how easy it’d be to develop a model deployment system using Python, thus avoiding a multi-tiered system. With the help of rpy2, it turns out that this wasn’t very difficult.
Continue Reading »
Posted in cheminformatics | Tagged cdk, python, qsar, R, REST, rpy2, solubility, web service | 1 Comment »
The folks at the EBI have been doing some great work on the CDK. A major effort is underway to revamp JChemPaint and part of this involves improving the rendering of 2D depictions. While not complete I rebuilt a version of the CDK 1.2.x branch with the latest rendering code from the jchempaint-primary branch and updated the CDK web service. The results are much nicer, though there’s scope for improvements. See for example
http://rguha.ath.cx/~rguha/cicc/rest/depict/c1ccccc1
http://rguha.ath.cx/~rguha/cicc/rest/depict/C1CCCCC12CCCCC2
http://rguha.ath.cx/~rguha/cicc/rest/depict/CC(=O)OC1=CC=CC=C1C(=O)O
http://rguha.ath.cx/~rguha/cicc/rest/depict/c1ccccc1CC=CC%23N
Thanks to Gilleain and Egon for pointing me in the right direction. Anybody using this service should see the new depictions automatically
Posted in software, Uncategorized | Tagged cdk, depiction | Leave a Comment »
The current version of the REST interface to the CDK descriptors allowed one to access descriptor values for a SMILES string by simply appending it to an URL, resulting in something like
http://rguha.ath.cx/~rguha/cicc/rest/desc/descriptors/ org.openscience.cdk.qsar.descriptors.molecular.ALOGPDescriptor/c1ccccc1COCC
This type of URL is pretty handy to construct by hand. However, as Pat Walters pointed out in the comments to that post, SMILES containing ‘#’ will cause problems since that character is a URL fragment identifier. Furthermore, the presence of a ‘/’ in a SMILES string necessitates some processing in the service to recognize it as part of the SMILES, rather than a URL path separator. While the service could handle these (at the expense of messy code) it turned out that there were subtle bugs.
Based on Pats’ suggestion I converted the service to use base64 encoded SMILES, which let me simplify the code and remove the bugs. As a result, one cannot append the SMILES directly to the URL’s. Instead the above URL would be rewritten in the form
http://rguha.ath.cx/~rguha/cicc/rest/desc/descriptors/ org.openscience.cdk.qsar.descriptors.molecular.ALOGPDescriptor/YzFjY2NjYzFDT0ND
All the example URL’s described in my previous post that involve SMILES strings, should be rewritten using base64 encoded SMILES. So to get a document listing all descriptors for “c1ccccc1COCC” one would write
http://rguha.ath.cx/~rguha/cicc/rest/desc/descriptors/YzFjY2NjYzFDT0ND
and then follow the links therein.
While this makes it a little harder to directly write out these URL’s by hand, I expect that most uses of this service would be programmatic – in which case getting base64 encoded SMILES is trivial.
Posted in cheminformatics, software | Tagged base64, descriptors, python, REST, web service | 3 Comments »
As part of my work at IU I have been implementing a number of cheminformatics web services. Initially these were SOAP, but I realized that REST interfaces make life much easier. (also see here) As a result, a number of these services have simple REST interfaces. One such service provides molecular descriptor calculations, using the CDK as the backend. Thus by visiting (i.e., making a HTTP GET request) a URL of the form
http://rguha.ath.cx/~rguha/cicc/rest/desc/descriptors/CC(=O)
you get a simple XML document containing a list of URL’s. Each URL represents a specific “resource”. In this context, the resource is the descriptor values for the given molecule. Thus by visiting
http://rguha.ath.cx/~rguha/cicc/rest/desc/descriptors/ org.openscience.cdk.qsar.descriptors.molecular.ALOGPDescriptor/CC(=O)C
one gets another simple XML document that lists the names and values of the AlogP descriptor. In this case, the CDK implementation evaluates AlogP, AlogP2 and molar refractivity – so there are actually three descriptor values. On the other hand something like the molecular weight descriptor gives a single value. To just see the list of available descriptors visit
http://www.chembiogrid.org/cheminfo/rest/desc/descriptors
which gives an XML document containing a series of links. Visiting one of these links gives the “descriptor specification” – information on the vendor, version, reference to a descriptor ontology and so on.
(I should point out that the descriptors available in this service are from a pretty old version of the CDK. I really should update the descriptors to the 1.2.x versions)
Applications
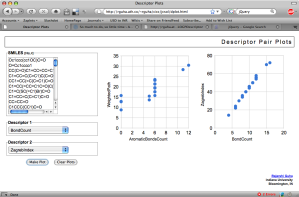
This type of interface makes it easy to whip up various applications. One example is the PCA analysis of compound collections. Another one I put together today based on a conversation with Jean-Claude was a simple application to plot pairs of descriptor values for a collection of SMILES.
The app is pretty simple (and quite slow, since it uses synchronous GET’s to the descriptor service for each SMILES and has to make two calls for each SMILES – hey, it was a quick hack!). Currently, it’s a bit restrictive – if a descriptor calculates multiple values, it will only use the first value. To see how many values a molecular descriptor calculates, see the list here.
With a little more effort one could easily have a pretty nice online descriptor calculation application rivaling a standalone application such as the the CDK descriptor GUI
Also,if you struggle with nice CSS layouts, the CSS Layout Collection is a fantastic resource. And jQuery rocks.
Posted in cheminformatics, software | Tagged cdk, descriptor, google, javascript, REST, web service | 4 Comments »